B. Shrestha, A. Vertes
Analytical Chemistry, 2014, 86, 4308–4315
DOI: dx.doi.org/10.1021/ac500007t | Impact factor (2011):
DOWNNLOAD | ONLINE
SUMMARY
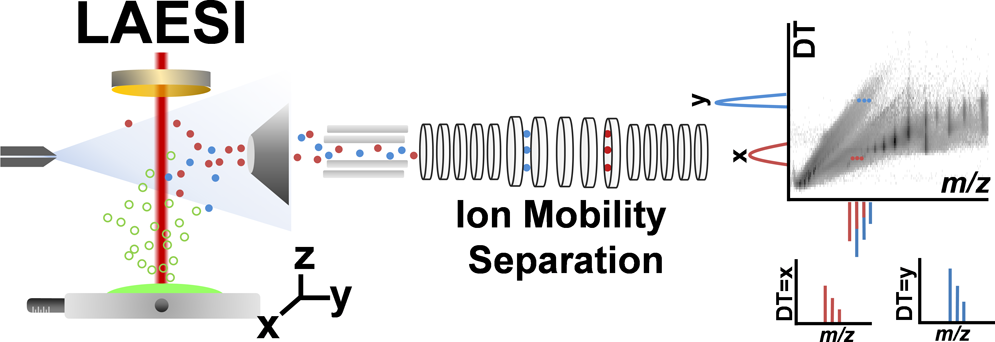
Ambient ionization methods, such as laser ablation electrospray ionization (LAESI), facilitate the direct analysis of unperturbed cells and tissues in their native states.However, the lack of a separation step in these ionization techniques results in limited molecular coverage due to interferences, ion suppression effects, and the lack of ability to differentiate between structural isomers and isobaric species. In this contribution, LAESI mass spectrometry (MS) coupled with ion mobility separation (IMS) is utilized for the direct analysis of protein mixtures, megakaryoblast cell pellets, mouse brain sections, and Arabidopsis thaliana leaves. We demonstrate that the collision cross sections of ions generated by LAESI are similar to the ones obtained by ESI. In various applications, LAESI-IMS-MS allows for the high-throughput separation and mass spectrometric detection of biomolecules on the millisecond time scale with enhanced molecular coverage. For example, direct analysis of mouse brain tissue without IMS had yielded ∼300 ionic species, whereas with IMS over 1 100 different ions were detected. Differentiating between ions of similar mass-to-charge ratios with dissimilar drift times in complex biological samples removes some systematic distortions in isotope distribution patterns and improves the fidelity of molecular identification. Coupling IMS with LAESI-MS also expands the dynamic range by increasing the signal-to-noise ratio due to the separation of isobaric or other interfering ionic species. We have also shown that identification of potential biomarkers by LAESI can be enhanced by using the drift times of individual ions as an additional parameter in supervised orthogonal projections to latent structures discriminant analysis. Comparative analysis of drift time versus mass-to-charge ratio plots was performed for similar tissue samples to pinpoint significant metabolic differences.